Specify a linear model to compare 2 groups
To identify differentially expressed genes for the leukemia experiment, you need to translate the following linear model to R:
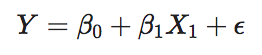
where \(X_{1}\) is equal to 1 for progressive cancers and 0 for stable cancers (note: R automatically chooses the base condition by alphabetical order).
Bu egzersiz
Differential Expression Analysis with limma in R
kursunun bir parçasıdırEgzersiz talimatları
The ExpressionSet object eset with the leukemia data has been loaded in your workspace.
- Use
model.matrixto construct a design matrix with an intercept coefficient and a coefficient that indicates the disease status.
Uygulamalı interaktif egzersiz
Bu örnek kodu tamamlayarak bu egzersizi bitirin.
# Create design matrix for leukemia study
design <- ___(~___, data = ___(eset))
# Count the number of samples modeled by each coefficient
colSums(design)