Spezifiziere ein lineares Modell zum Vergleich von 2 Gruppen
Um differentiell exprimierte Gene im Leukämie-Experiment zu identifizieren, musst du das folgende lineare Modell nach R übertragen:
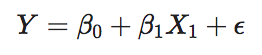
wobei \(X_{1}\) für progressive Krebsfälle 1 und für stabile Krebsfälle 0 ist (Hinweis: R wählt die Basisbedingung automatisch in alphabetischer Reihenfolge).
Diese Übung ist Teil des Kurses
Differenzielle Expressionsanalyse mit limma in R
Anleitung zur Übung
Das ExpressionSet-Objekt eset mit den Leukämie-Daten wurde in deinen Arbeitsbereich geladen.
- Verwende
model.matrix, um eine Designmatrix mit einem Achsenabschnitt (Intercept) und einem Koeffizienten zu erstellen, der den Krankheitsstatus angibt.
Interaktive Übung
Vervollständige den Beispielcode, um diese Übung erfolgreich abzuschließen.
# Create design matrix for leukemia study
design <- ___(~___, data = ___(eset))
# Count the number of samples modeled by each coefficient
colSums(design)