Théorie DGE : métadonnées
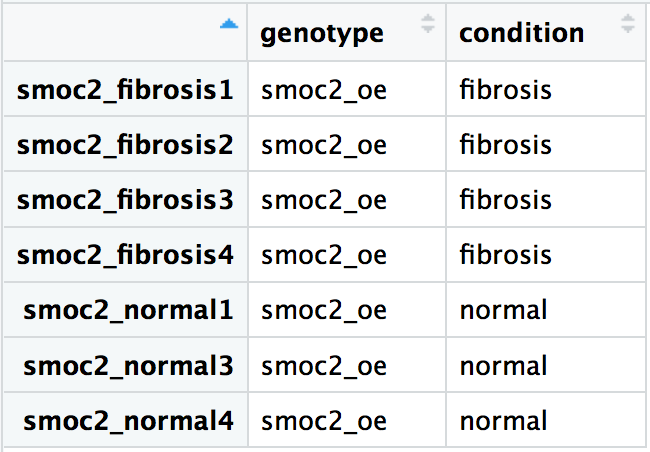
Utilisez les informations ci-dessous pour créer une table de métadonnées pour les comptages de fibrose appelée metadata, avec les colonnes genotype et condition. Les noms d’échantillons (par ex. smoc2_fibrosis1, smoc2_fibrosis2, etc.) doivent être les noms de lignes de la table :
Cet exercice fait partie du cours
RNA-Seq avec Bioconductor en R
Instructions
- Créez un vecteur de caractères nommé
genotypepour les données ci-dessus en utilisantc(). - Créez un vecteur de caractères nommé
conditionpour les données ci-dessus en utilisantc(). - Créez une table de données nommée
smoc2_metadataen utilisantdata.frame()et les vecteurs de caractèresgenotypeetcondition. - Créez un vecteur de noms d’échantillons avec
c()et affectez-le aux noms de lignes de la table à l’aide derownames().
Exercice interactif pratique
Essayez cet exercice en complétant cet exemple de code.
# Create genotype vector
genotype <- c(___)
# Create condition vector
condition <- c(___)
# Create data frame
smoc2_metadata <- data.frame(___, ___)
# Assign the row names of the data frame
rownames(smoc2_metadata) <- c(___)