DGE Theory: Metadata
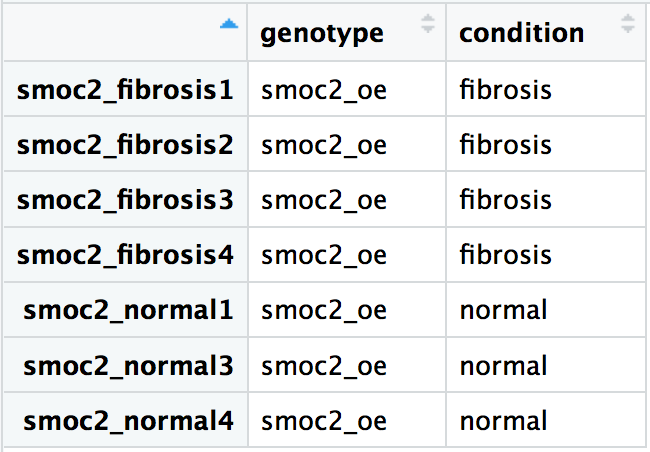
Use the information below to create a metadata data frame for the fibrosis count data called metadata with columns genotype and condition. The sample names (e.g. smoc2_fibrosis1, smoc2_fibrosis2, etc.) should be the row names of the data frame:
Deze oefening maakt deel uit van de cursus
RNA-Seq with Bioconductor in R
Oefeninstructies
- Create a character vector called
genotypefor the above data usingc(). - Create a character vector called
conditionfor the above data usingc(). - Create a data frame called
smoc2_metadatausingdata.frame()and thegenotypeandconditioncharacter vectors. - Create a vector of sample names using
c()and assign it to the row names of the data frame usingrownames().
Praktische interactieve oefening
Probeer deze oefening eens door deze voorbeeldcode in te vullen.
# Create genotype vector
genotype <- c(___)
# Create condition vector
condition <- c(___)
# Create data frame
smoc2_metadata <- data.frame(___, ___)
# Assign the row names of the data frame
rownames(smoc2_metadata) <- c(___)