Plot other views
Any two dimensions of an array can form an image, and slicing along different axes can provide a useful perspective. However, unequal sampling rates can create distorted images.
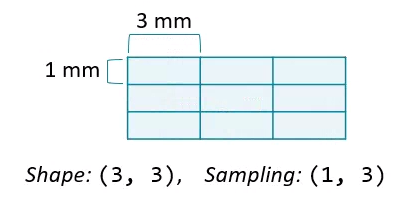
Changing the aspect ratio can address this by increasing the width of one of the dimensions.
For this exercise, plot images that slice along the second and third dimensions of vol. Explicitly set the aspect ratio to generate undistorted images.
Diese Übung ist Teil des Kurses
Biomedical Image Analysis in Python
Anleitung zur Übung
- Slice a 2D plane from
volwhere "axis 1" is256. - Slice a 2D plane from
volwhere "axis 2" is256. - For each image, calculate the aspect ratio by dividing the image "sampling" rate for axis 0 by its opponent axis. This information is in
vol.meta. - Plot the images in a subplots array. Specify the
aspectratio for each image, and setcmap='gray'.
Interaktive Übung
Vervollständige den Beispielcode, um diese Übung erfolgreich abzuschließen.
# Select frame from "vol"
im1 = vol[:, 256, :]
im2 = ____
# Compute aspect ratios
d0, d1, d2 = ____
asp1 = d0 / d2
asp2 = ____
# Plot the images on a subplots array
fig, axes = plt.subplots(nrows=2, ncols=1)
axes[0].imshow(im1, cmap='gray', ____)
____
plt.show()