Criando o objeto DE
OBSERVAÇÃO: Carregar este exercício pode levar um pouco mais de tempo.
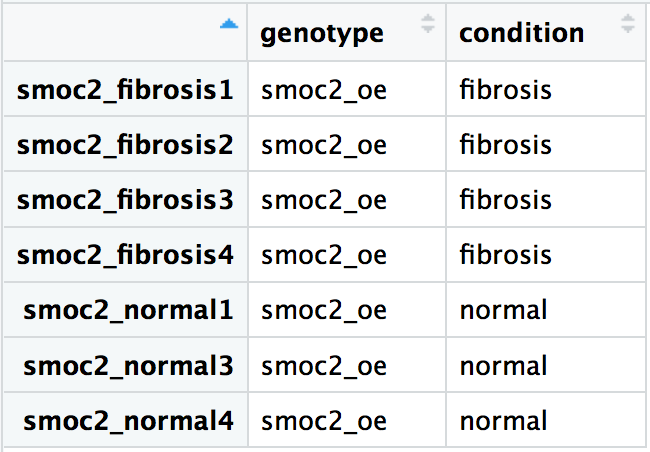
Usando nossas amostras com superexpressão de smoc2, crie o objeto do DESeq2 de modo que a fórmula de design especifique a comparação das diferenças de expressão entre as amostras de fibrose e normais. Os metadados do experimento estão abaixo. Os dados já foram lidos com as amostras na mesma ordem para as contagens brutas de smoc2, reordered_smoc2_rawcounts, e para os metadados, smoc2_metadata.
Este exercício faz parte do curso
RNA-Seq com Bioconductor em R
Instruções do exercício
Crie um objeto DESeq2 chamado
dds_smoc2usando a funçãoDESeqDataSetFromMatrix(), especificando os argumentos:countData,colDataedesign.Execute a função
DESeq()para estimar os fatores de tamanho, calcular as dispersões e realizar o ajuste e o teste do modelo.
Exercício interativo prático
Experimente este exercício completando este código de exemplo.
# Create DESeq2 object
dds_smoc2 <- ___(___ = ___,
___ = ___,
___ = ~ condition)
# Run the DESeq2 analysis
___ <- ___(___)