Filter and wrapper methods
Questions about reducing the dimensionality of a dataset are highly common in machine learning interviews. One way to reduce the dimensionality of a dataset is by only selecting relevant features in your dataset.
Here you'll practice a filter method on the diabetes DataFrame followed by 2 different styles of wrapper methods that include cross-validation. You will be using pandas, matplotlib.pyplot and seaborn to visualize correlations, process your data and apply feature selection techniques to your dataset.
The feature matrix with the dropped target variable column (progression) is loaded as X, while the target variable is loaded as y.
Note that pandas, matplotlib.pyplot, and seaborn have already been imported to your workspace and aliased as pd, plt, and sns respectively.
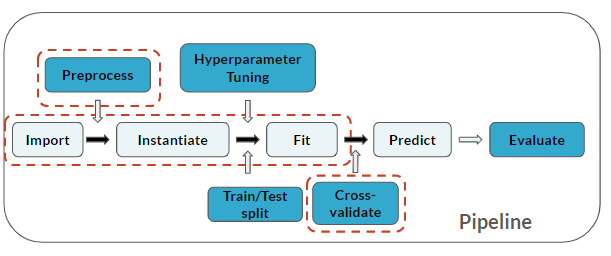
Notice you've added a Cross-validate step to your pipeline (which applies to the last 3 steps):
This exercise is part of the course
Practicing Machine Learning Interview Questions in Python
Hands-on interactive exercise
Have a go at this exercise by completing this sample code.
# Create correlation matrix and print it
cor = ____.____()
print(____)
# Correlation matrix heatmap
plt.figure()
sns.____(____, annot=True, cmap=plt.cm.Reds)
plt.show()
# Correlation with output variable
cor_target = abs(cor["progression"])
# Selecting highly correlated features
best_features = ____[____ > ____]
print(____)