Crear el objeto DE
NOTA: Cargar este ejercicio puede tardar un poco más.
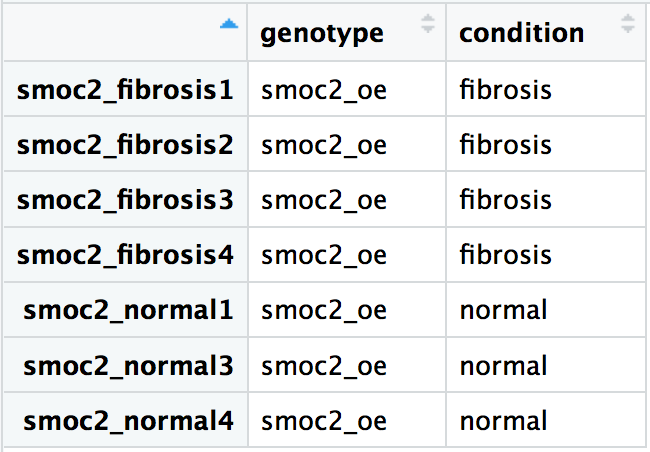
Usando nuestras muestras con sobreexpresión de smoc2, crea el objeto de DESeq2 de forma que la fórmula de diseño especifique la comparación de las diferencias de expresión entre las muestras de fibrosis y las normales. Los metadatos del experimento se muestran abajo. Ya hemos leído los datos con las muestras en el mismo orden tanto para los conteos brutos de smoc2, reordered_smoc2_rawcounts, como para los metadatos, smoc2_metadata.
Este ejercicio forma parte del curso
RNA-Seq con Bioconductor en R
Instrucciones del ejercicio
Crea un objeto DESeq2 llamado
dds_smoc2usando la funciónDESeqDataSetFromMatrix()especificando los argumentos:countData,colDataydesign.Ejecuta la función
DESeq()para estimar los factores de tamaño, calcular las dispersiones y realizar el ajuste del modelo y las pruebas.
Ejercicio interactivo práctico
Prueba este ejercicio y completa el código de muestra.
# Create DESeq2 object
dds_smoc2 <- ___(___ = ___,
___ = ___,
___ = ~ condition)
# Run the DESeq2 analysis
___ <- ___(___)