Deduce MNAR
In the previous exercise, you worked on identifying the type of missing values given the missingness summary. In this exercise, you'll continue on that spree to affirmatively identify data that is Missing Not at Random (MNAR).
The missingness summary for the diabetes DataFrame is as below.
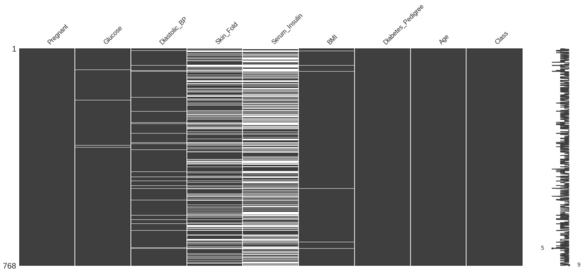
Your goal is to sort the diabetes DataFrame on Serum_Insulin and identify the correlation between Skin_Fold and Serum_Insulin.
Note that we've used a proprietary display() function instead of plt.show() to make it easier for you to view the output.
Diese Übung ist Teil des Kurses
Dealing with Missing Data in Python
Anleitung zur Übung
- Import the
missingnopackage asmsno. - Sort the values of
Serum_Insulincolumn indiabetes. - Visualize the missingness summary of
Serum_Insulinwithmsno.matrix().
Interaktive Übung
Vervollständige den Beispielcode, um diese Übung erfolgreich abzuschließen.
# Import missingno as msno
___
# Sort diabetes dataframe on 'Serum Insulin'
sorted_values = ___.___(___)
# Visualize the missingness summary of sorted
___.___(___)
# Display nullity matrix
display("/usr/local/share/datasets/matrix_sorted.png")